Modeling
This section covers modeling completing data query part and pre-modeling. We will reach out models fitting and ensembling
Model fitting
Model fitting is the ability we give to a machine to generalize data on which the model is trained. In practice, it consists of provide training data and some parameters to models in a way that can approach the test data the model don’t see yet. See this section to learn more about data partition into training and test data.
Firstly, select the models that you plan to use. We recommend selecting multiple models and fit them to check which provide the best performance. The choice of model is done in pre-modeling part of nimo clicking on ![]() button. In the modal that appears, click in the algorithm field to choose the model(s) you want to perform.
button. In the modal that appears, click in the algorithm field to choose the model(s) you want to perform.
For example purpose, we select Random Forest (RF), Support Vector Machine (SVM) and Generalized Linear Models (GLM) algorithm. Then click on Ok button to close the modal. This is required if you exit the app after select the algorithm(s) and set up a working directory. Except that you don’t need to define a working directory again. The interface for model fitting appear now in Fitting menu of nimo app.

You need the input data. If a break was taken until Extraction and data stemming from was saved in local, you must import it checking ‘Use existing data’ check box. If no, the data got from extraction will automatically integrated. For example purpose, we import data previously processed and available here. After import data, an option relevant to Ensemble of Small Models (ESM) appears to let decide if it is standard model that will be fitted or ESM model.
Our occurrence data is poor, although for many years (from 2016 to 2022). The sample size is limited to 25 records. So we prefer fit an Ensemble Small Models, checking ESM option. This switches model fitting fields from standard to ESM.

As you can remark, Random forest field disappear. This algorithm is not available for ESM. In case predictors don’t appear for selection, uncheck and check ‘Use existing data’ check box. Now let’s move on to explain some of the common and distinct parameters of these two models
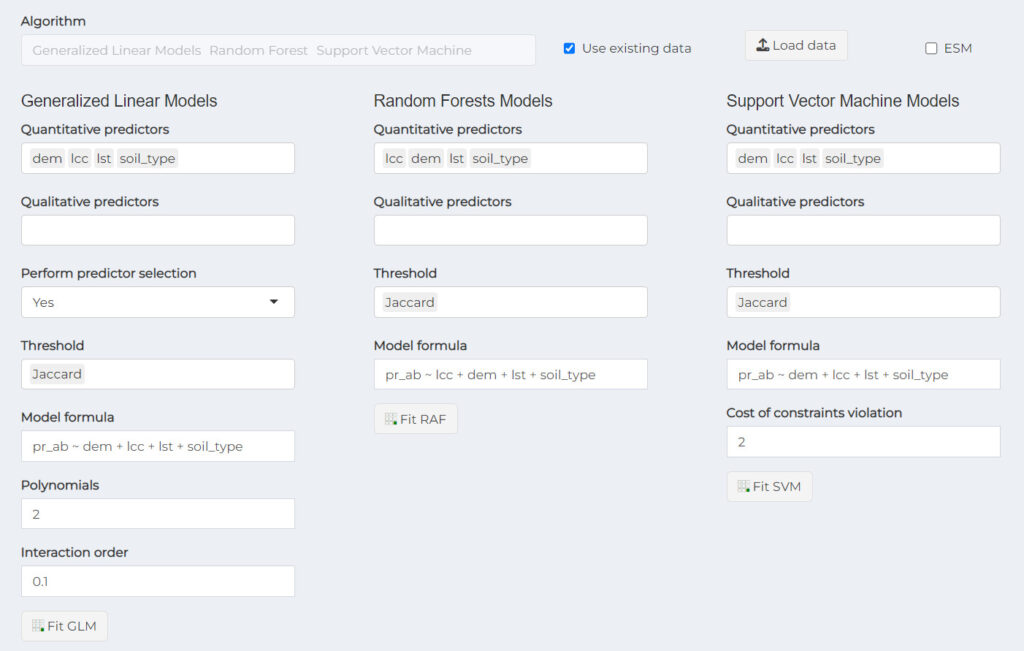
Quantitative predictors: column names of quantitative predictor variables. This can only construct models with continuous variables and does not allow categorical variables.
Threshold: used to get binary suitability values (i.e. 0,1). It is useful for threshold-dependent performance metrics. It is possible to use more than one threshold type. All threshold available are used if any threshold is provided explicitly. The following threshold criteria are available:
- Sensitivity = specificity: Threshold at which the sensitivity and specificity are equal.
- TSS: Threshold at which the sum of the sensitivity and specificity is the highest (also known as threshold that maximizes the TSS).
- Jaccard : The threshold at which Jaccard is the highest.
- Sorensen: The threshold at which Sorensen is highest.
- FPB: The threshold at which FPB (F-measure on Presence-Background data) is highest.
- Sensitivity: Threshold based on a specified sensitivity value.
Polynomials – GLM: If used with values >= 2 model will use polynomials for those continuous variables (i.e. used in predictors argument). The fact that ESM are constructed with few occurrences, polynomials can cause overfitting.
Interaction order – GLM: The interaction order between explanatory variables. Default is 0. Because ESM are constructed with few occurrences it is recommended not to use interaction terms.
Cost of constraints violation – SVM: Cost of constraints violation, the ‘C’ constant of the regularization term in the Lagrange formulation.
Each model has Fit … at bottom to run fitting. Those button launch a modal that shows the modal output, like Model’s summary, Performance metric and Predicted suitability.
🔄All process above are same to fit standard models, except some additional parameters.
Small Model summaries
We provide an example interpretation of models output
Standard models fitting
In this section, we will discover how to fit standard models and ensemble them. The algorithms used in Ensemble Small Models previously are renews. Proceed to algorithms selection. Be sure ESM option is unchecked in fitting menu because we don’t want fit ESM here.
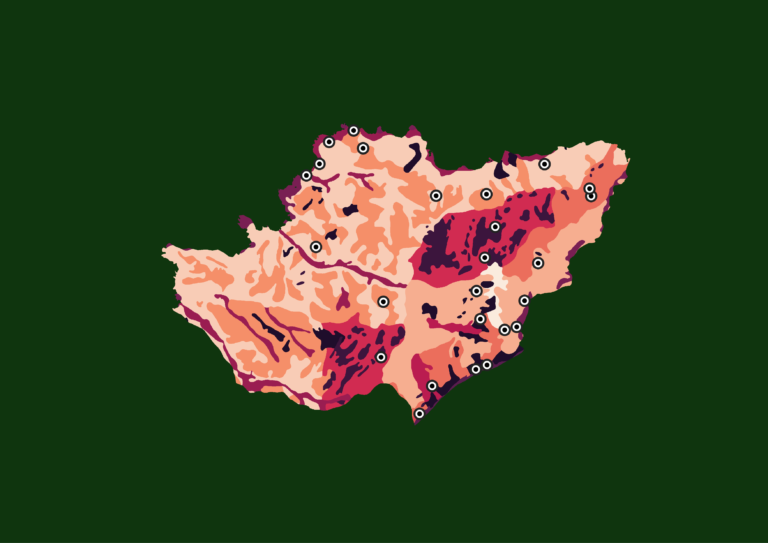
The output is similar to previous model’s summaries. For GLM, If you select Yes to perform predictor selection, predictors will be selected based on backward step-wise approach. The selected predictors are represented by the model with lower AIC, indicating that this model provides a reasonably good fit to the data. Look the explanation results of the Random Forest classification model used for species distribution modeling of Aardvarks in Pendjari National Park in pane below.
Ensemble models
Ensemble techniques are methods that combine the predictions of multiple individual models (often called base models or weak learners) to improve overall predictive performance. The key idea behind ensemble methods is to harness the collective intelligence of multiple models to make more accurate and robust predictions than any individual model could achieve on its own. To ensemble models in nimo, go in Ensemble menu.
The models we just fit are listed. We can select Support Vector Machine (SVM) and Generalized Linear Models (GLM) and ![]() it. The resulted model can be used for prediction, as we will see in post-modeling.
it. The resulted model can be used for prediction, as we will see in post-modeling.