Welcome to Nimo's support
Through this resource, we will give all necessaries explanations to understand nimo‘s standard and how it works. Four (04) main parts will be reach.
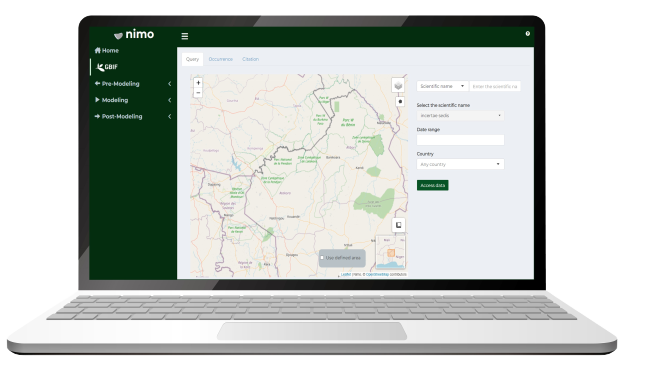
Query Occurence data
Via GBIF API, you will learn how to access occurrence data and inject it into species distribution modelling workflow.
Pre-modelling
Classically, there are some practice to set species distribution modelling environment ready. We will show you how can you do that by preparing modeling input data (e.g., species occurrences thinning, sample pseudo-absences or background points, delimitation of calibration area).
Modelling
This section groups actions related to modeling construction and validation with default hyperparameter values and models searching for the best hyperparameter values combination. Furthermore, you understand how to construct and validate an Ensemble of Small Models.
Post-modelling
We will explore the tools related to models’ geographical predictions, evaluation, and correction.
Install nimo
First thing first, install nimo R package that is not yet on CRAN. The development version can be installed from github. Copy and paste R code chunk bellow and run it in RStudio (IDE of R) or directly in R. You can get these sofwares from official website of RStudio here if you don’t have yet.
⚠️ NOTE: The version 1.4-22 of terra package is causing errors when trying to install flexsdm and so nimo. Please, first install a version ≥ 1.5-12 of terra package available on CRAN or development version of terra and then nimo.
# Install pak if it is not already installed
if (!require("pak", character.only = TRUE)) {
install.packages("pak")
}
pak::pkg_install("stangandaho/nimo")
Launch the app
Once nimo is installed, load the package and call nimo() function.
library(nimo)
nimo()The button below link each part of the app as explained above. All necessaries concept and part of nimo are explained with screenshot and example.
